Report Displayers Examples
Report displayers you can use in your own Mine and some examples created for specific data types in modMine, FlyMine and metabolicMine.
The following displayers can all be used for data loaded by standard InterMine parsers. To see how to configure them check out FlyMine's webconfig-model.xml.
SequenceFeature summary#
Applicable for any SequenceFeature. It shows length, sequence export, chromosome location, cyto location and SO term (where present).

Protein sequence#
Applicable for Protein. It shows length, sequence export.
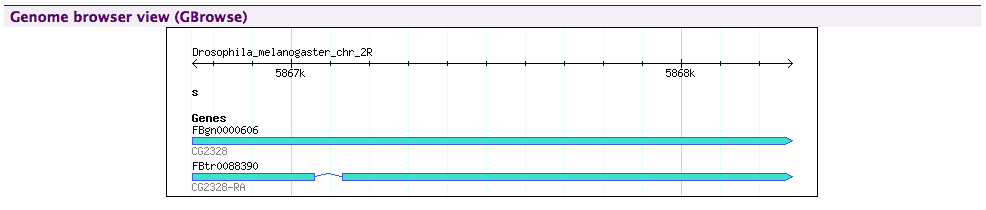
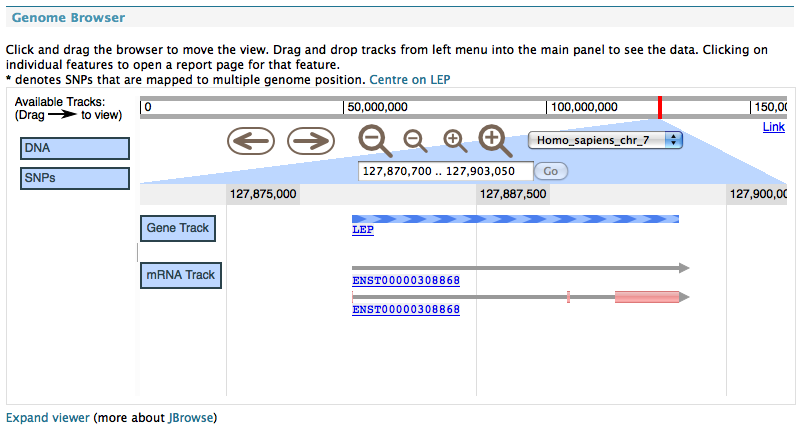
GBrowse#
Shows an inline image from a configured GBrowse instance.
This also needs two properties to be configured in the minename.properties file: gbrowse.prefix and gbrowse_image.prefix which give the location of a running GBrowse instance.
Homologues#
Shows a table of organism and homologous genes of homologues per organism.
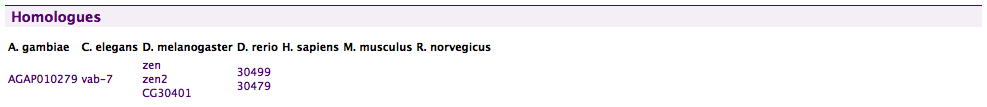
Note that FlyMine includes a specific displayer to show the twelve Drosophila species as a phylogenetic tree.
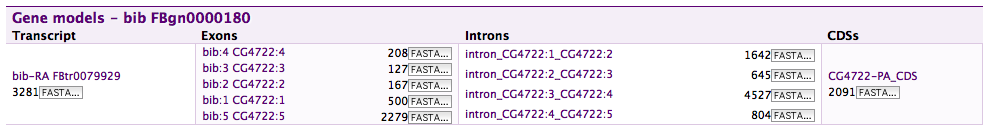
Gene structure#
Displays transcripts, exons, introns, UTRs and CDSs if present in the model and for the particular organism. It can be added to report pages for any of these feature types and will find the parent gene and show all transcripts, highlighting the feature of the actual report page.
Gene Ontology#
Simple display of GO terms and evidence codes for a gene, grouped by branch in the ontology. It groups by the three main ontologies (function, process and component), so you may need to run the GO source.

UniProt comments#
A clear view of curated comments from UniProt (SwissProt) applied to a protein, or for a gene will show comments from all proteins of the gene.

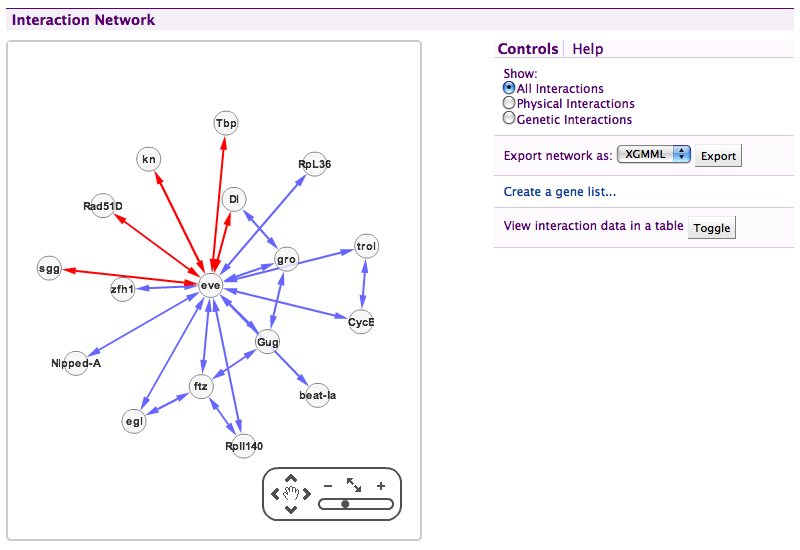
Interaction network#
Uses the Cytoscape Web plugin to display physical and genetics interactions. The interaction displayer links to report pages, allows creation of a gene list of the whole network and can show tabular interaction data. Read NetworkDisplayer for details.
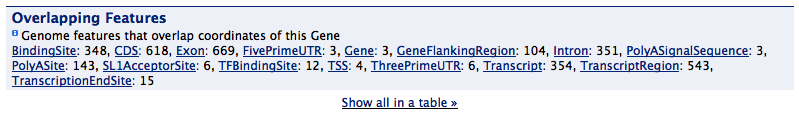
Overlapping features#
A summary view of features that overlap the chromosome location of the reported feature. If the gene structure displayer is also used, it will exclude any features that are part of the same gene model i.e. it won't report that a gene overlaps it's own exons.
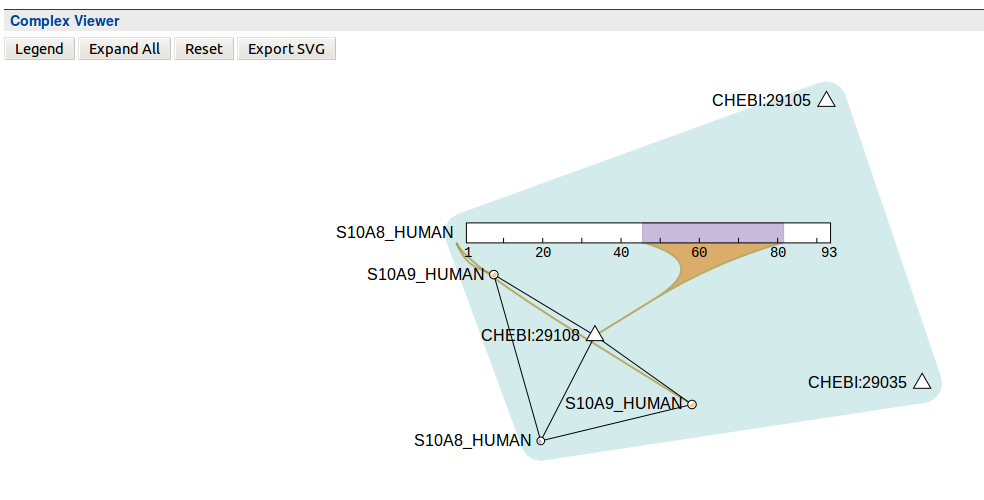
Complexes - Protein interactions#
Viewer displaying complex interactions. Data must be loaded from IntAct. Original Source: http://interactionviewer.org/.
Specific Displayers#
There are some displayers created for specific data sets in FlyMine, metabolicMine or modMine that may not be re-usable in other Mines but could be adapted or provide inspiration.
 JavaScript library in FlyMine.](/im-docs/assets/images/drosophila_homology_displayer-d6669402f6aac868d495e95b5c9987ac.png)
 JavaScript library.](/im-docs/assets/images/modMine-heatmap-ccd61514d7b592555d4bb575a6cb5851.png)
