InterMine
Welcome to InterMine! 👋#
Before diving into details on the InterMine Server documentation and its capabilities, let us introduce some background information about the InterMine software.
Disparate data in, unified data out#
InterMine is a data warehouse system that integrates biological data sources, making it easy to query and analyse data. It is an open-source project that is available for use under the open-source LGPL license. InterMine was developed by the Micklem lab at the University of Cambridge with the Wellcome Trust's support; complementary projects have been funded by the National Human Genome Research Institute (NIH/NHGRI) and the Biotechnology and Biological Sciences Research Council (BBSRC). InterMine includes an attractive, user-friendly interface, known as BlueGenes, that works ‘out of the box’ and can be easily customised for your specific needs, as well as a powerful, scriptable web-service API to allow programmatic access to your data.
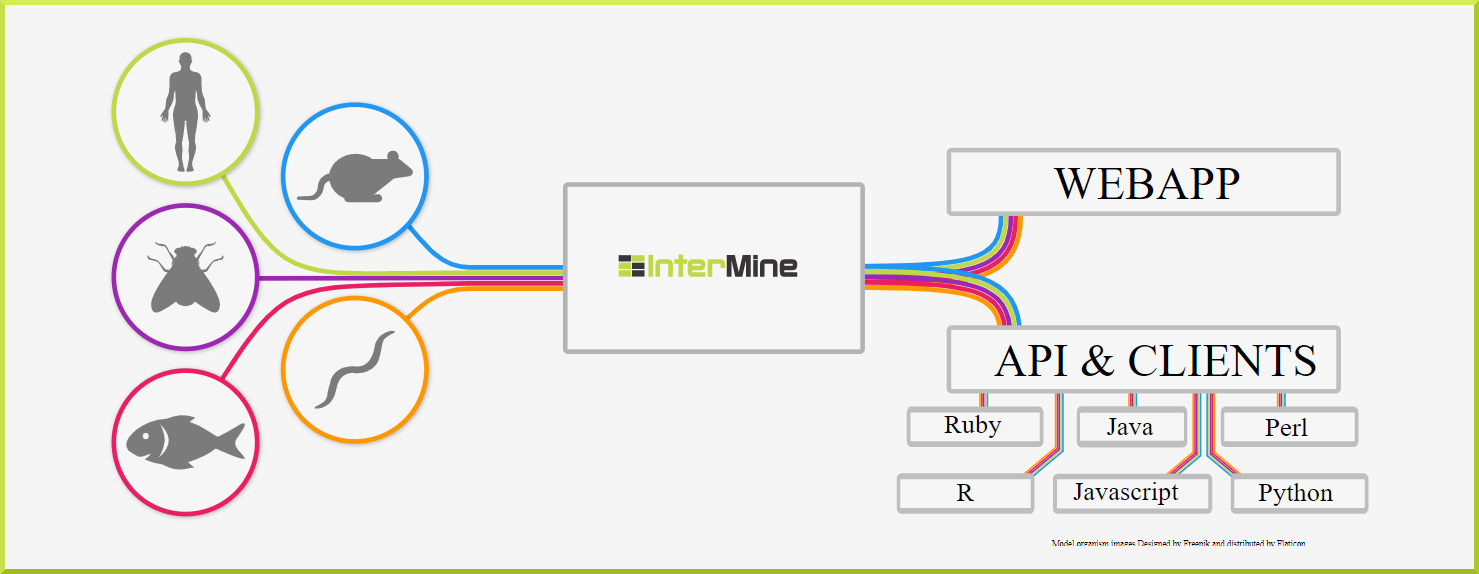
There's an InterMine for (almost) anyone!#
There are many different InterMines worldwide, covering a broad range of model organisms and life science research areas, including:
- FlyMine - a data warehouse of integrated fruit fly genetic, genomic and proteomic data
- HumanMine - an integrated database of Homo sapiens genomic data
- MouseMine - an integrated data warehouse of mouse genomic data, developed by MGI
- YeastMine - an integrated data warehouse of yeast genomic data, developed by SGD
- ZebrafishMine - an integrated data warehouse of zebrafish genomic data, developed by ZFIN
- RatMine - an integrated data warehouse of rat genomic data, developed by RGD
- TargetMine - a data warehouse for candidate gene prioritisation and drug target discovery, developed at NIBIO, Japan.
- ThaleMine - a data warehouse for Arabidopsis thaliana Col-0 for the ARAPORT project
- PhytoMine - an integrated data warehouse of over 50 plant genomes from Phytozome.
For the full list of InterMines, please see the registry.
