InterMine User Documentation
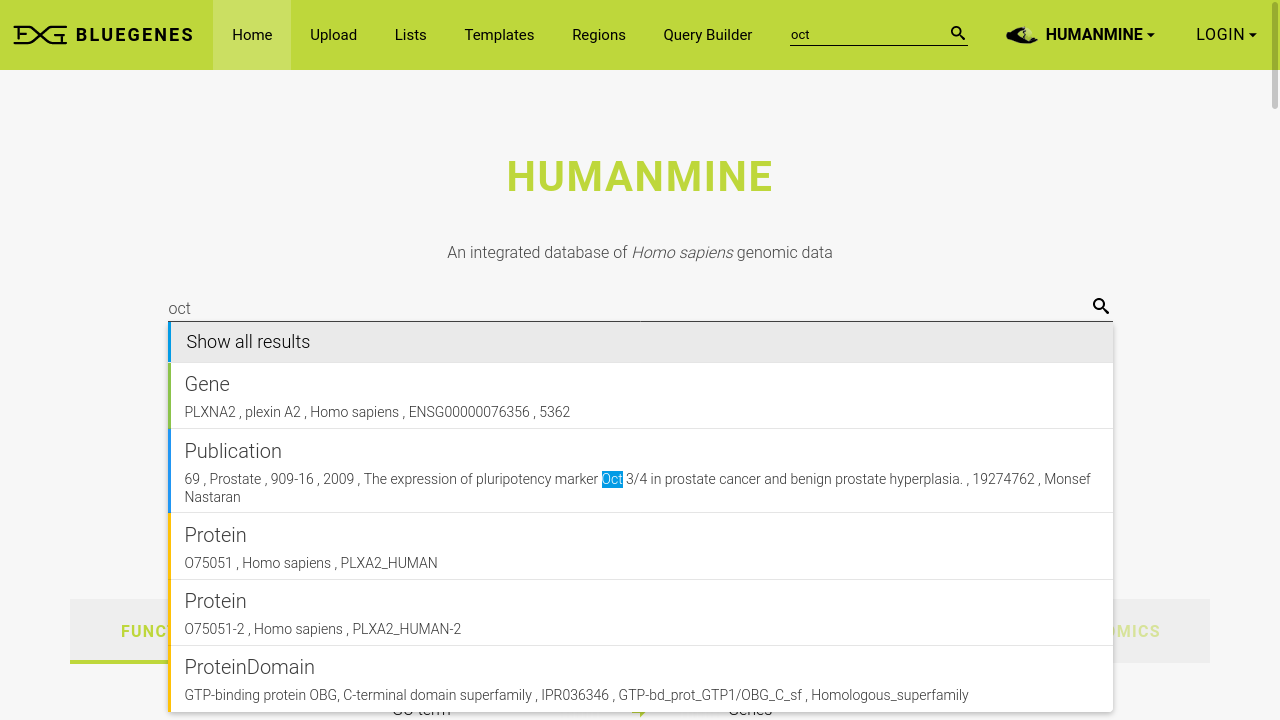
Explore various biological sources
InterMine integrates heterogeneous biological data sources, often but not always focused around a specific organism. The user interface allows you to explore the rich variety of data on a single platform, with only a short click away from exporting code for InterMine API clients when you wish to take a programmatic approach.
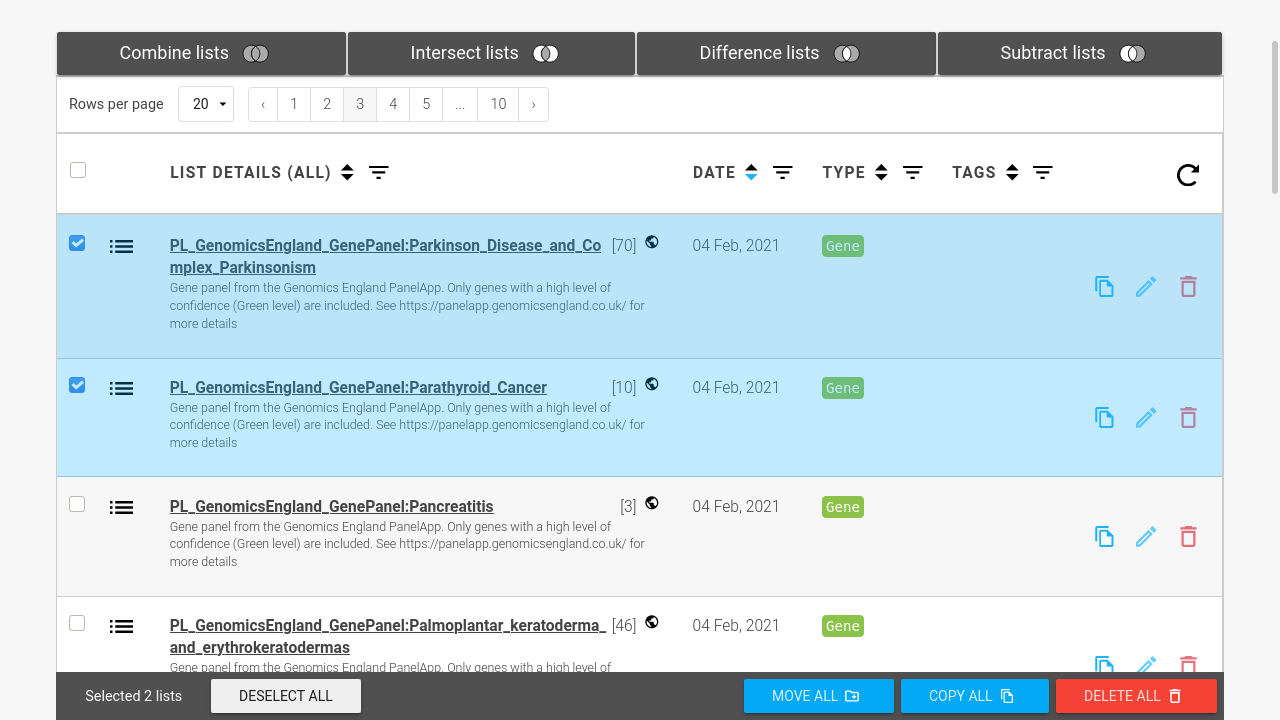
Create lists of genes, SNPs, publications, etc.
Objects in InterMine are standardised and distinct entities mapped to a biological subject. This allows you to create lists of entities (such as gene or protein lists). Lists can be analysed using set operations (such as union and intersect) and used in batch queries. Lists can be created from queries in an InterMine or uploaded from an external source.
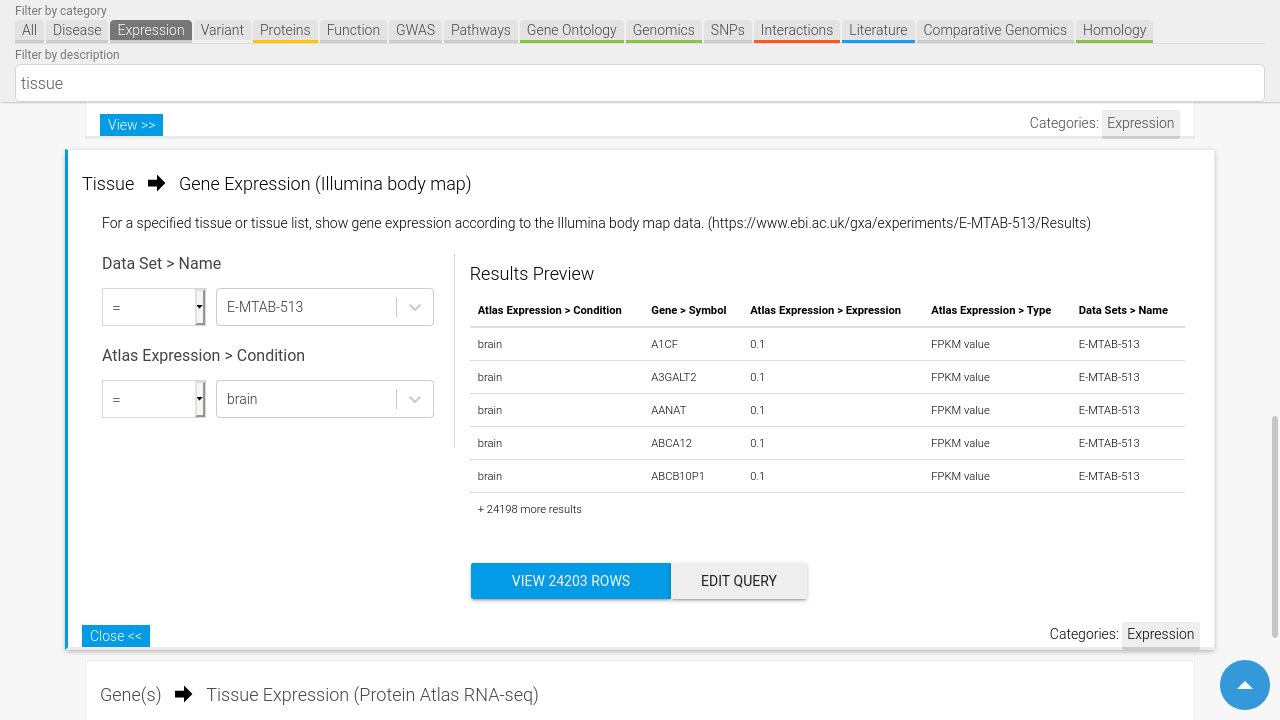
Run predefined queries
The maintainers of an InterMine make available useful queries by predefining them as templates. These contain fields that you can edit such as specifying a specific gene or tissue for expression data. Advanced users are also able to create their own queries by utilising the powerful Query Builder interface.
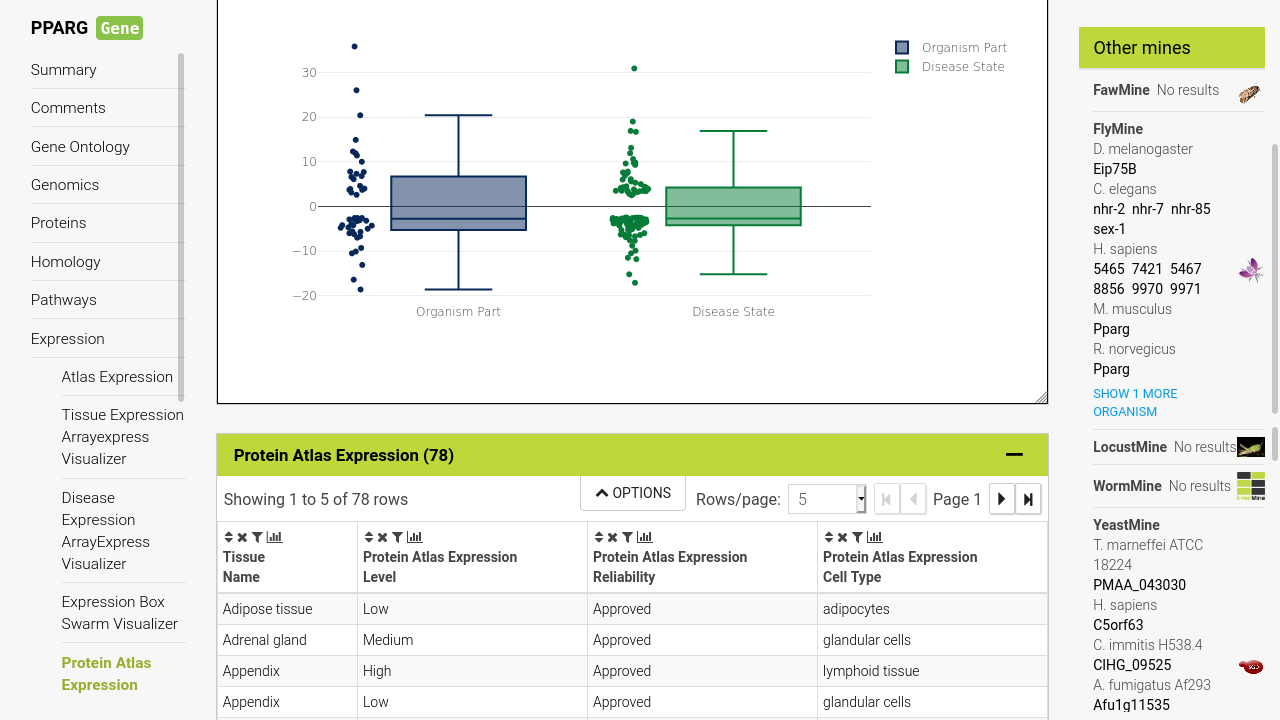
Analyse objects through rich reports
When you wish to focus on one specific object, such as a gene, protein or publication - you can view their report to see all data related to them, in addition to custom visualisations and compatible predefined queries. Any data in a table can be clicked to navigate to its respective report.